|
Motif analysis
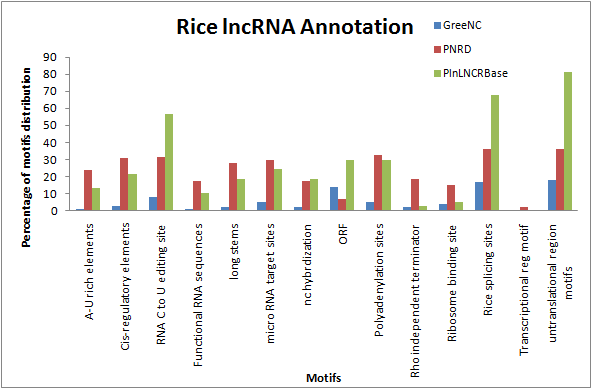
Fig.2 - From the comparison of 14 types of motifs present in 6064 rice lncRNA sequences from the GreeNC, PNRD and PLNlncRBase databases, untranslated region (UTR) and rice splicing site motifs occurred maximum in all the databases. lncRNA sequences mostly contained UTR, Rice splicing sites, RNA C to U editing site, Polyadenylation site, ORF, miRNA target sites, CIS regulatory element motifs. 210 types of snoRNA were found to have ncRNA hybridization Transcriptional regulatory motifs were least or not present in the lncRNA sequences. Repeat analysis showed the presence of transposable repeat elements in lncRNA sequences.
|

